98% green spaghetti, sliced and chopped
- 2018/15/04
- 18 min read
This is the latest stop in an analysis tour of free-range menu data.
One of the goals of fishing for real recipes is to be able to suss out patterns in how foods are combined and in what amounts in order to be able to generate new recipes. However, this post will mostly eschew creating anything useful and just mess around with the words in recipes themselves.
As a step toward creating new ingredients and recipes in interesting ways, we’ll
tag ingredient words with their parts of speech to find the most common noun-adjective pairs, and
create some menu mad libs.
For a sneak peek, here are a few that were created while I was putting this together.
| ingredient |
|---|
| 1/8 small, black round oil leaves - extract into cups |
| 3 cherry ribs, fully lime |
| 1 olive boneless, broken |
| 1/2 extra Sour carrots, lime for fluid pieces |
| 1/3 frozen onion, virgin |
| 1 Beef oil |
| 6 orange Mashed fresh Ranch |
| 1 Lindsay® |
So that deliciousness is what we have to look forward to at the end of this little experiment. Yummo.
Bit of Background
Up to this point we’ve scraped recipes from Allrecipes.com and tidied the resulting data, extracted ingredient unit names, and matched them their corresponding portion_abbreviations. We also munged the free text numbers into usable portion amounts (multiplying and averaging units where necessary to create) and converted these units into grams.
A glance at what the result of that looks like:
set.seed(123)
recipes_df %>%
sample_n(10) %>%
select(recipe_name, ingredients, portion_abbrev, portion, converted) %>%
map_dfc(replace_na, "") %>%
kable()| recipe_name | ingredients | portion_abbrev | portion | converted |
|---|---|---|---|---|
| African Chicken in Spicy Red Sauce | 1 1/2 cups chopped onion | cup | 1.5 | 354.88 |
| Homemade Seasoned Salt | 1/4 cup kosher salt | cup | 0.25 | 59.15 |
| Calico Rice | 2 tablespoons picante sauce or salsa | tbsp | 2 | 29.57 |
| Poached Beef Fillet Served with its Own Broth and Baby Winter Vegetables | 8 ounces oxtail or beef shin | oz | 8 | 226.8 |
| Easy Puebla-Style Chicken Mole | 3 cups fat-free, low-sodium chicken broth | cup | 3 | 709.76 |
| Pork Picante | 1/4 teaspoon cayenne pepper | tsp | 0.25 | 1.23 |
| Candy Cane Cookies from Gold Medal® Flour | 2 large egg yolks | 2 | ||
| Chocolate-Raspberry Muffins | 1 1/2 teaspoons baking powder | tsp | 1.5 | 7.39 |
| Bhindi Masala (Spicy Okra Curry) | 1 teaspoon salt | tsp | 1 | 4.93 |
| Spaghetti and Meatballs (Paleo Style) | 1/2 cup cooked quinoa | cup | 0.5 | 118.29 |
Tagging Parts of Speech
In order to be able to combine ingredients in ways that might make sense, it’ll be useful to know the parts of speech of each word that makes up a given ingredient. The goal now is going to be to tag each word with its part of speech while retaining our tidy data format.
I’ll use the RDRPOSTagger package (beware, it does depend on rJava which can be a pain to install) to assign a part of speech (POS) to every word in every recipe.
First we define a model to use as our POS tagger.
library(RDRPOSTagger)
tagger <- rdr_model(language = "English", annotation = "POS")Let’s see how the main rdr_pos() function works by tagging a few ingredients. The first argument is the model, which for us is tagger.
tagger %>%
rdr_pos(recipes_df$ingredients[1:3]) %>%
as_tibble() %>%
kable()| doc_id | token_id | token | pos |
|---|---|---|---|
| d1 | 1 | 1 | CD |
| d1 | 2 | (12 | CD |
| d1 | 3 | inch) | NN |
| d1 | 4 | pre | NN |
| d1 | 5 | - | : |
| d1 | 6 | baked | JJ |
| d1 | 7 | pizza | NN |
| d1 | 8 | crust | NN |
| d2 | 1 | 1 | CD |
| d2 | 2 | 1/2 | CD |
| d2 | 3 | cups | NNS |
| d2 | 4 | shredded | JJ |
| d2 | 5 | mozzarella | NNP |
| d2 | 6 | cheese | NN |
| d3 | 1 | 1 | CD |
| d3 | 2 | (14 | CD |
| d3 | 3 | ounce) | NN |
| d3 | 4 | jar | NN |
| d3 | 5 | pizza | NN |
| d3 | 6 | sauce | NN |
The pos column shows the acronym for the token’s part of speech.
A POS acronym-to-description key is below1:
| Tag | Description |
|---|---|
| CC | Coordinating conjunction |
| CD | Cardinal number |
| DT | Determiner |
| EX | Existential there |
| FW | Foreign word |
| IN | Preposition or subordinating conjunction |
| JJ | Adjective |
| JJR | Adjective, comparative |
| JJS | Adjective, superlative |
| LS | List item marker |
| MD | Modal |
| NN | Noun, singular or mass |
| NNS | Noun, plural |
| NNP | Proper noun, singular |
| NNPS | Proper noun, plural |
| PDT | Predeterminer |
| POS | Possessive ending |
| PRP | Personal pronoun |
| PRP$ | Possessive pronoun |
| RB | Adverb |
| RBR | Adverb, comparative |
| RBS | Adverb, superlative |
| RP | Particle |
| SYM | Symbol |
| TO | to |
| UH | Interjection |
| VB | Verb, base form |
| VBD | Verb, past tense |
| VBG | Verb, gerund or present participle |
| VBN | Verb, past participle |
| VBP | Verb, non-3rd person singular present |
| VBZ | Verb, 3rd person singular present |
| WDT | Wh-determiner |
| WP | Wh-pronoun |
| WP$ | Possessive wh-pronoun |
| WRB | Wh-adverb |
Modifying the Tagging Function
The rdr_pos() function gets us most of the way there, but I’ll make a slight adjustment to it in order to preserve more explicitly the relationship between the input and output.
By default, rdr_pos() names each doc_id as the default paste("d", seq_along(x), sep = "") where x is the input vector. That’s how we get d1, d2, etc. Instead of this default, we can ask this function to act more like a mutate by setting doc_id equal to our input’s recipe name concatenated with its ingredient. Then we only need to do a separate() on the <break> to dislodge our recipe name from its ingredient column. This explicitly relates input to output.
We’ll also want to include the rest of our original dataframe in the output. To keep things more compact, we nest() our POS tags into a data column so that we have one row per input before left_join()ing on our original dataframe.
recipe_sample <- recipes_df %>%
slice(1:5)
tagged_sample <- tagger %>%
RDRPOSTagger::rdr_pos(recipe_sample$ingredients,
doc_id = paste(recipe_sample$recipe_name,
recipe_sample$ingredients,
sep = "<break>")) %>%
separate(doc_id, into = c("recipe_name", "ingredients"), sep = "<break>") %>%
nest(-recipe_name, -ingredients) %>%
left_join(recipe_sample)The result is easily unnested. I’ll show just a few columns here for space’s sake.
tagged_sample %>%
unnest() %>%
slice(1:5) %>%
select(recipe_name, ingredients, token, pos, portion) %>%
kable()| recipe_name | ingredients | token | pos | portion |
|---|---|---|---|---|
| Johnsonville® Three Cheese Italian Style Chicken Sausage Skillet Pizza | 1 (12 inch) pre-baked pizza crust | 1 | CD | 12 |
| Johnsonville® Three Cheese Italian Style Chicken Sausage Skillet Pizza | 1 (12 inch) pre-baked pizza crust | (12 | CD | 12 |
| Johnsonville® Three Cheese Italian Style Chicken Sausage Skillet Pizza | 1 (12 inch) pre-baked pizza crust | inch) | NN | 12 |
| Johnsonville® Three Cheese Italian Style Chicken Sausage Skillet Pizza | 1 (12 inch) pre-baked pizza crust | pre | NN | 12 |
| Johnsonville® Three Cheese Italian Style Chicken Sausage Skillet Pizza | 1 (12 inch) pre-baked pizza crust | - | : | 12 |
It’s worth noting that some punctuation (like commas and periods) gets tagged as is, and some (like dashes and semicolons) gets a POS of ":". I’m not totally sure what the logic is behind that is, but it means that dashes, semicolons, and colons are treated interchangeably, whereas commas and periods are their own unique POS.
tagger %>%
rdr_pos("foo , . - ; bar") %>%
kable()| doc_id | token_id | token | pos |
|---|---|---|---|
| d1 | 1 | foo | NN |
| d1 | 2 | , | , |
| d1 | 3 | . | . |
| d1 | 4 | - | : |
| d1 | 5 | ; | : |
| d1 | 6 | bar | NN |
If the distinction between dashes and semicolons is important, then when making mad libs we may down the line want to and only sub in punctuation marks for ones that are lexically equivalent.
Tagging a Dataframe
Let’s throw that bit of work into a function.
In tag_df() we’ll also get rid of all parentheses that often surround digits in order to be able to combine them with other words without any stray open or closed parentheses cluttering things up.
tag_df <- function(df, model = tagger) {
df <- df %>%
mutate(
ingredients = ingredients %>% map_chr(str_replace_all, "[\\(\\)]", "")
)
inp <- df$ingredients
tagged <- model %>%
rdr_pos(inp,
doc_id = glue::glue("{df$recipe_name}<break>{df$ingredients}")) %>%
separate(doc_id,
into = c("recipe_name", "ingredients"),
sep = "<break>") %>%
nest(-recipe_name, -ingredients) %>%
left_join(df) %>%
unnest()
return(tagged)
}And now we can tag our dataframe.
tagged_recipes_df <- recipes_df %>%
tag_df()What unique parts of speech do we have?
tibble("Tag" = unique(tagged_recipes_df$pos)) %>%
left_join(pos_table, by = "Tag") %>%
kable()| Tag | Description |
|---|---|
| CD | Cardinal number |
| NN | Noun, singular or mass |
| : | NA |
| JJ | Adjective |
| NNS | Noun, plural |
| NNP | Proper noun, singular |
| , | NA |
| . | NA |
| VBN | Verb, past participle |
| VB | Verb, base form |
| CC | Coordinating conjunction |
| MD | Modal |
| VBZ | Verb, 3rd person singular present |
| IN | Preposition or subordinating conjunction |
| JJR | Adjective, comparative |
| RB | Adverb |
| TO | to |
| VBG | Verb, gerund or present participle |
| DT | Determiner |
| VBD | Verb, past tense |
| NNPS | Proper noun, plural |
| VBP | Verb, non-3rd person singular present |
| ’’ | NA |
| JJS | Adjective, superlative |
| RBR | Adverb, comparative |
| PRP$ | Possessive pronoun |
| SYM | Symbol |
| RP | Particle |
Common Consecutive Word Pairs
Now that we have each word’s part of speech, so now let’s see how words within ingredients relate to one another.
If we take a dplyr::lag() of our tagged dataframe, we can see the relationship between words that come directly after one another. token comes first, and token_lag second.
We can read the token_lag and token columns left to right now to see each pair of words in the sequence they appear in in the recipe.
tagged_recipes_df %>%
group_by(recipe_name, ingredients) %>%
mutate(
token_id_lag = lag(token_id),
token_lag = lag(token),
pos_lag = lag(pos)
) %>%
ungroup() %>%
select(ingredients, token_lag, token, pos_lag, pos) %>%
map_df(replace_na, "") %>%
ungroup() %>%
slice(1:5) %>%
kable()| ingredients | token_lag | token | pos_lag | pos |
|---|---|---|---|---|
| 1 12 inch pre-baked pizza crust | 1 | CD | ||
| 1 12 inch pre-baked pizza crust | 1 | 12 | CD | CD |
| 1 12 inch pre-baked pizza crust | 12 | inch | CD | NN |
| 1 12 inch pre-baked pizza crust | inch | pre | NN | NN |
| 1 12 inch pre-baked pizza crust | pre | - | NN | : |
We can throw this into a function that allows us to filter to any specific combination of token and token_lag parts of speech. By default we’ll set both to "NN" for nouns. (We use %in% in the filter() instead of == so that we can supply multiple possibilities for the parts of speech to keep.)
find_pos_pairs <- function(df, pos_lag_keep = 'NN', pos_keep = 'NN') {
out <- df %>%
group_by(recipe_name) %>%
mutate(
token_id_lag = lag(token_id),
token_lag = lag(token),
pos_lag = lag(pos)
) %>%
select(
ends_with("id"), ends_with("lag"), everything()
) %>%
filter(pos %in% pos_keep & pos_lag %in% pos_lag_keep)
return(out)
}Using find_pos_pairs(), we can take untagged dataframe, tag it, and filter it a straightforward pipeline like this.
recipes_df %>%
tag_df() %>%
find_pos_pairs()If our dataframe is already tagged, we can skip the tag_df() step, pass Go, and collect our POS pairs.
Let’s do that by looking at adjectives that precede nouns.
adj_noun_pos <- tagged_recipes_df %>%
find_pos_pairs(pos_lag_keep = c("JJ", "JJR", "JJS"), # All the adj types
pos_keep = c("NN", "NNS", "NNP", "NNPS")) # All the noun typesNow a simple count of each distinct pair to see what the most common adj-noun combos are. These can be read left-to-right in the order they appear in ingredients.
adj_noun_pos_counts <- adj_noun_pos %>%
group_by(token_lag, token) %>%
count(sort = TRUE) %>%
rename(n_pairs = n) %>%
map_df(str_replace_all, "[\\()]", "") # Remove parens
adj_noun_pos_counts %>%
ungroup() %>%
slice(1:15) %>%
kable()| token_lag | token | n_pairs |
|---|---|---|
| black | pepper | 76 |
| brown | sugar | 38 |
| white | sugar | 34 |
| garlic | powder | 22 |
| Cheddar | cheese | 21 |
| red | pepper | 20 |
| virgin | olive | 19 |
| sour | cream | 18 |
| green | onions | 17 |
| fluid | ounce | 16 |
| fresh | basil | 16 |
| lean | ground | 16 |
| green | bell | 15 |
| red | onion | 15 |
| fresh | cilantro | 13 |
From these counts we can draw up a quick scatterplot of the most frequent adjective-noun pairs in our recipes.
ggplot(adj_noun_pos_counts %>% ungroup() %>%
top_n(15)) +
geom_point(aes(reorder(token, n_pairs), reorder(token_lag, n_pairs),
size = n_pairs), stat = "identity") +
ggtitle("Frequent Adjective-Noun Pairs") +
labs(x = "Noun", y = "Adjective", size = "N Pairs") +
theme_minimal(base_family = "Source Sans Pro") +
theme(axis.text.x = element_text(angle = 45, hjust = 1))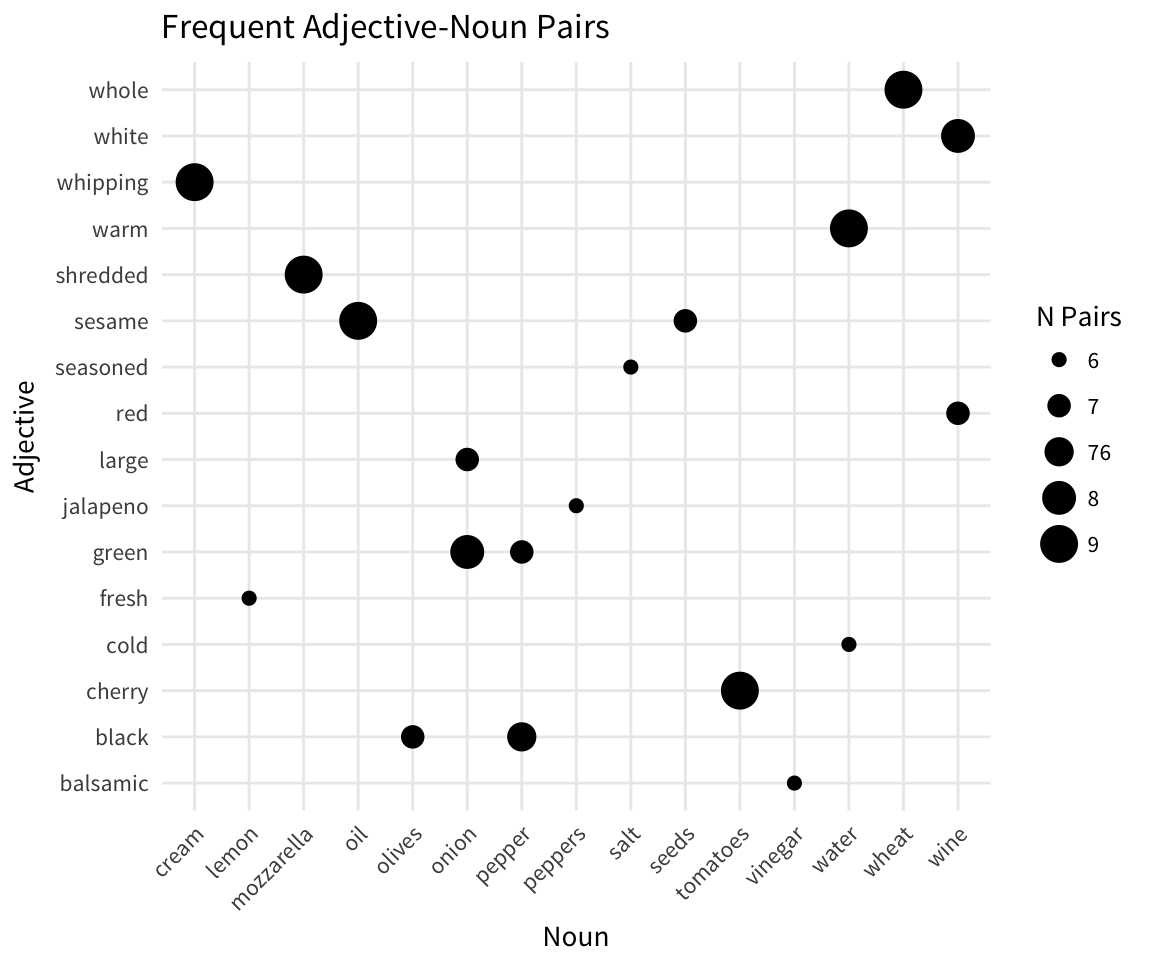
Source table was scraped and tidied into this dataframe.↩
That’s not as dramatic of a speedup as we would have gotten if we hadn’t reduced the amount of copies that need to be made at each iteration by allocating space in output vector jumping into the loop (using
out <- vector(mode = "character", length = n_libs)).↩
